Molecular dynamics (MD) simulations have long been a powerful tool for understanding the structural and dynamic properties of biomolecules at the atomic level. combining MD simulation and deep learning provide researchers with detailed insights into molecular interactions, conformational changes, and even the mechanisms underlying biological functions. However, as computational resources and experimental data continue to expand, the challenge of efficiently analyzing and extracting meaningful insights from these simulations has grown. This is where deep learning, with its ability to model complex relationships and patterns in large datasets, comes into play.
Combining MD simulation and deep learning is opening new avenues for advancing biomolecular research, enabling more accurate predictions and faster computational workflows. However, this integration also comes with challenges that need to be addressed. In this article, we will explore both the challenges and opportunities in combining MD simulation with deep learning, focusing on how this fusion can drive new biomolecular insights.
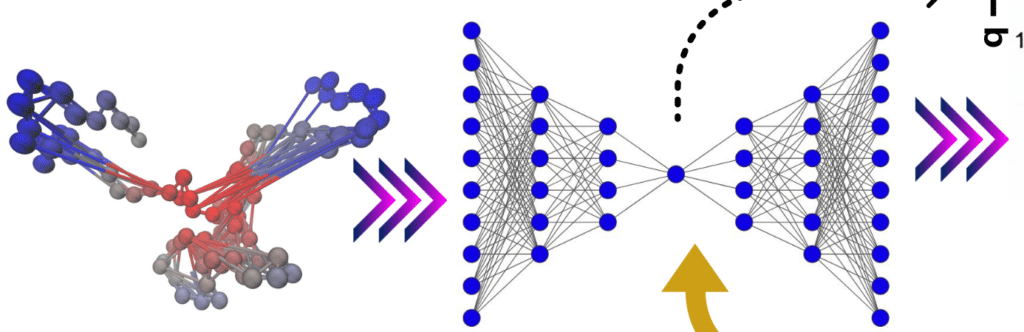
Overview of Molecular Dynamics Simulations
Molecular dynamics (MD) simulations model the behavior of molecules over time by solving Newton’s equations of motion. MD simulations provide high-resolution data about the physical movements of atoms and molecules, making them useful for studying various biological systems, including proteins, nucleic acids, and membranes. These simulations are commonly used to explore:
- Protein folding: Understanding how proteins adopt their functional three-dimensional structures.
- Drug-receptor interactions: Simulating how small molecules bind to proteins and other macromolecules.
- Conformational changes: Observing how molecules undergo structural changes under different conditions.
Despite their detailed output, MD simulations are computationally expensive and generate vast amounts of data, making it challenging to analyze and interpret results efficiently. As MD simulation datasets grow in size and complexity, traditional analysis methods may fall short, which is why integrating deep learning has become a viable solution.
The Role of Deep Learning in Biomolecular Simulations
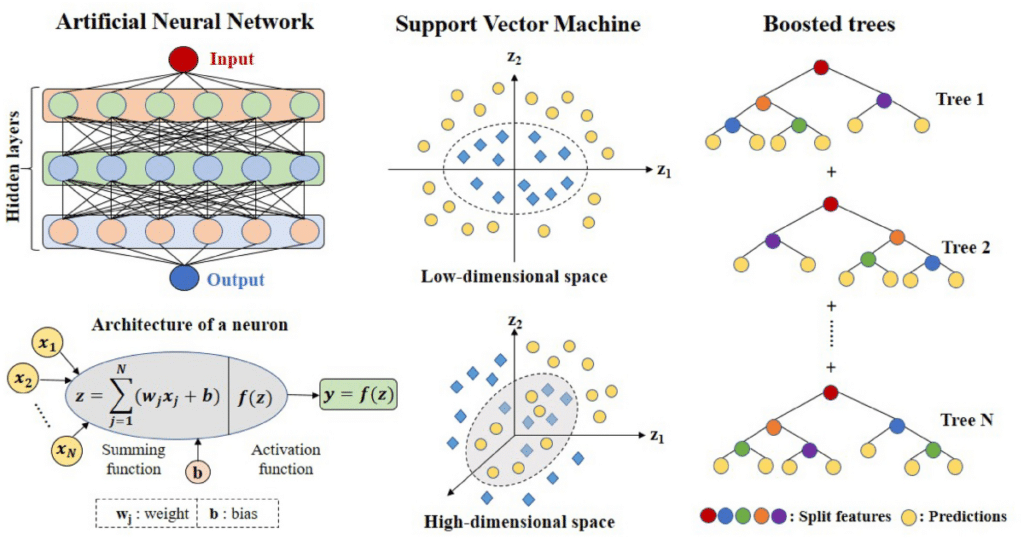
Deep learning, a subset of machine learning, excels at identifying patterns in large, high-dimensional datasets. It leverages neural networks, which can be trained to perform tasks like classification, regression, and pattern recognition. For MD simulations, deep learning algorithms can be applied to analyze trajectories, predict molecular behavior, and even generate new molecular structures.
Some of the primary benefits of integrating deep learning into MD simulations include:
- Automated Feature Extraction: Deep learning models can automatically identify relevant features from complex molecular trajectories, saving time and effort compared to manual feature selection.
- Predictive Modeling: Deep learning algorithms can be trained on large-scale MD simulation data to predict molecular properties or interactions that have not been explicitly simulated.
- Dimensionality Reduction: High-dimensional data from MD simulations can be reduced into lower-dimensional representations, making it easier to visualize and analyze.
- Speed: Once trained, deep learning models can provide fast predictions, accelerating the analysis process of MD simulations, which is often computationally intensive.
While the potential benefits of combining MD simulation and deep learning are significant, several challenges remain in realizing the full potential of this integration.
Challenges in Combining MD Simulation and Deep Learning
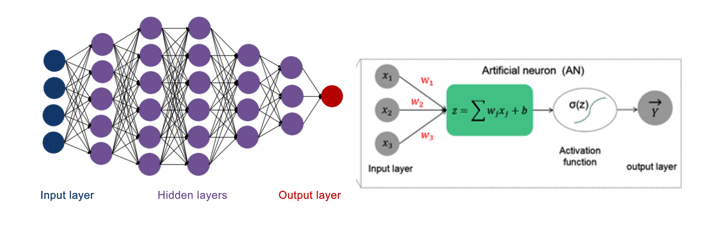
1. Data Availability and Quality
One of the primary challenges in using deep learning for MD simulations is the availability and quality of training data. MD simulations generate massive amounts of data, but these datasets are often noisy or incomplete. Training deep learning models on noisy data can result in models that fail to generalize to new data or produce inaccurate predictions.
Additionally, the need for labeled data is a significant barrier. Supervised deep learning models require labeled datasets for training, which can be difficult to obtain in the context of molecular simulations. Manually labeling MD simulation data is time-consuming and often requires expert knowledge.
Solution: Using unsupervised or semi-supervised learning methods, which do not require extensive labeled data, can mitigate this challenge. Transfer learning, where a model is pre-trained on a large dataset and then fine-tuned on a smaller, domain-specific dataset, is another approach to addressing limited data.
2. Model Interpretability
Deep learning models, particularly neural networks, are often referred to as “black boxes” because they do not provide clear explanations for how they arrive at their predictions. For many researchers in molecular biology and chemistry, understanding the underlying mechanisms driving molecular behavior is as important as making accurate predictions. However, the opaque nature of deep learning models can make it difficult to extract interpretable biomolecular insights from them.
Solution: Explainable AI (XAI) methods, such as attention mechanisms and saliency maps, can be integrated into deep learning models to highlight the regions of molecular simulations that are most relevant for predictions. These tools can help provide a better understanding of what the model is learning and offer insights into the underlying molecular mechanisms.
3. Computational Costs
Training deep learning models on large MD simulation datasets requires significant computational resources. While deep learning models can accelerate predictions once trained, the training phase itself can be computationally expensive, particularly for deep networks. This can limit the accessibility of deep learning to researchers without access to high-performance computing clusters or GPUs.
Solution: Advances in hardware, such as cloud-based GPU resources, and software optimization, such as the use of specialized libraries like TensorFlow and PyTorch, can help reduce the computational burden. Additionally, techniques like model compression and pruning can be used to reduce the size and complexity of deep learning models without sacrificing accuracy.
4. Generalization to New Systems
MD simulations are often system-specific, meaning that a model trained on one system (e.g., a particular protein or nucleic acid) may not generalize well to new systems. For deep learning models to be useful in biomolecular research, they must generalize to a wide variety of molecular systems without retraining from scratch for every new application.
Solution: Transfer learning can help address this challenge by enabling models trained on one molecular system to be fine-tuned for a new system using a smaller amount of additional data. This reduces the need for extensive retraining and makes the models more adaptable.
Opportunities in Combining MD Simulation and Deep Learning
Despite the challenges, the combining MD simulation and deep learning presents numerous exciting opportunities for advancing biomolecular research.
1. Accelerating Drug Discovery
Deep learning models can be used to predict how small molecules interact with target proteins, significantly accelerating the drug discovery process. By integrating deep learning into MD simulations, researchers can rapidly screen large libraries of compounds to identify potential drug candidates with high binding affinity to a target protein. This approach is already showing promise in the development of treatments for diseases such as cancer, neurodegenerative disorders, and infectious diseases.
2. Exploring Protein Folding
Deep learning models, such as AlphaFold, have demonstrated the ability to predict protein structures with high accuracy. By combining MD simulations with deep learning, researchers can gain new insights into protein folding mechanisms and study how misfolding contributes to diseases such as Alzheimer’s and Parkinson’s.
3. Understanding Conformational Dynamics
Deep learning models can be trained to identify specific conformational states of proteins or other biomolecules from MD simulation trajectories. These models can help researchers identify functionally relevant conformations that may be difficult to capture using traditional analysis methods.
4. Data-Driven Force Fields
In MD simulations, the accuracy of results depends on the quality of the force fields used to describe molecular interactions. Deep learning models offer the potential to develop data-driven force fields that are more accurate and generalizable than traditional force fields, leading to more reliable simulation results.
Conclusion
The integration of combining MD simulation and deep learning represents a new frontier in biomolecular research. While there are challenges to overcome, such as data quality, model interpretability, and computational costs, the opportunities are vast. By leveraging the strengths of both approaches, researchers can unlock new insights into the behavior of biomolecules, accelerate drug discovery, and gain a deeper understanding of complex biological systems.
As tools and techniques for combining MD simulations and deep learning continue to evolve, researchers can look forward to a future where these methods play a central role in uncovering the molecular mechanisms underlying health and disease.
If you want to explore more about applications of combining MD Simulation with Deep Learning you can join us in Dubai for an exciting 2.5 Day Masterclass. More information is available HERE
