In the field of genomics, understanding the behavior of genes is essential to unlocking the mysteries of biology and disease. Two important techniques, gene expression profiling and pathway enrichment analysis, help researchers unravel the complexity of genetic regulation, allowing for a deeper understanding of biological processes, disease mechanisms, and therapeutic targets. This article provides a beginner’s guide to gene expression profiling and pathway enrichment analysis, explaining their significance, methods, and applications.
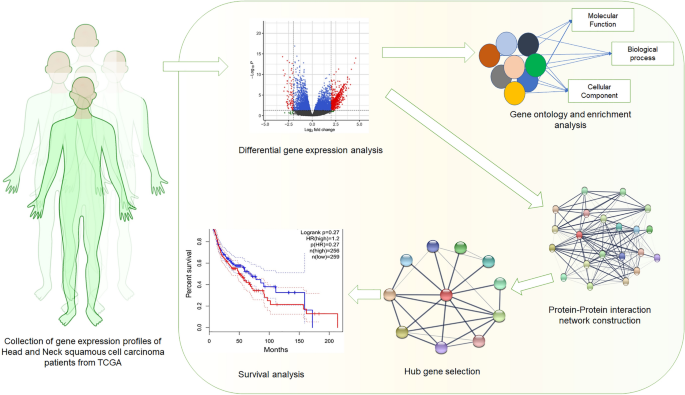
What is Gene Expression Profiling?
Gene expression profiling is a powerful technique used to measure the activity of thousands of genes simultaneously. This method allows researchers to understand which genes are “turned on” or “off” in a particular cell type or under specific conditions, such as in a disease state or response to treatment.
Key Concepts in Gene Expression Profiling
- Gene Expression: Refers to the process by which the information encoded in a gene is used to synthesize proteins or RNA molecules that play functional roles in the cell. The level of gene expression varies between different cell types, tissues, and in response to environmental or experimental conditions.
- Transcriptome: The complete set of RNA molecules produced in a cell or tissue at a particular time is called the transcriptome. Gene expression profiling allows scientists to capture this transcriptome to study the functional state of the cell.
- Gene Expression Profiling Technologies: There are several technologies used to perform gene expression profiling:
- Microarrays: An older method where pre-defined sequences are fixed on a chip, allowing scientists to measure the expression of known genes.
- RNA sequencing (RNA-Seq): A more modern and comprehensive approach that provides a high-resolution view of the transcriptome by sequencing all RNA molecules in a sample. RNA-Seq has become the preferred method for gene expression profiling due to its ability to detect novel transcripts and alternative splicing events.
The Workflow of Gene Expression Profiling
- Sample Collection: Biological samples, such as tissue or cell lines, are collected and the RNA is extracted for analysis.
- RNA Quantification: The quantity of RNA in each sample is measured to ensure accurate representation of gene activity.
- Library Preparation and Sequencing: For RNA-Seq, the extracted RNA is converted into complementary DNA (cDNA) and sequenced using high-throughput sequencing platforms.
- Data Analysis: After sequencing, bioinformatics tools are used to align the reads to a reference genome, quantify gene expression levels, and identify differentially expressed genes between conditions.
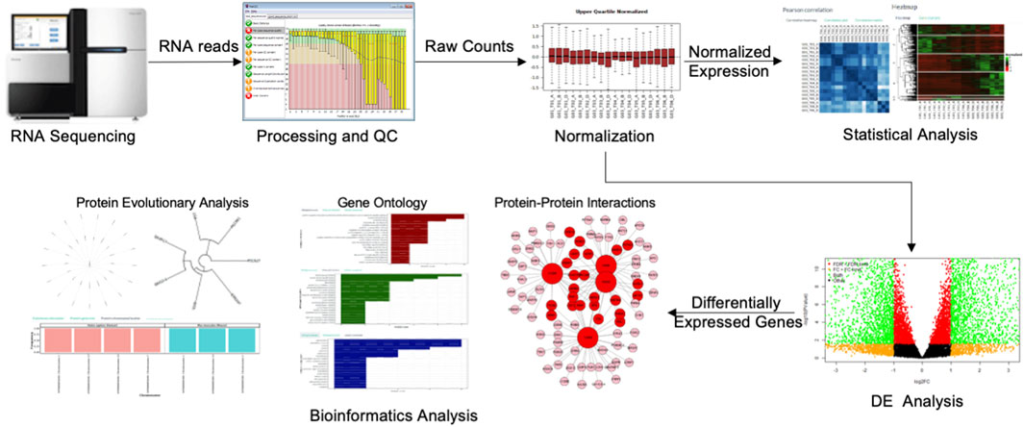
Applications of Gene Expression Profiling
Gene expression profiling has numerous applications in biology and medicine:
- Cancer Research: By comparing gene expression profiles of normal and tumor tissues, researchers can identify oncogenes and tumor suppressors.
- Drug Development: Profiling gene expression in response to drug treatments helps identify how drugs affect cellular pathways.
- Personalized Medicine: Understanding individual gene expression profiles can lead to more personalized treatment strategies, especially in cancer therapy.
- Developmental Biology: Studying gene expression changes during different stages of development helps uncover the molecular mechanisms driving organismal growth.
What is Pathway Enrichment Analysis?
While gene expression profiling tells us which genes are expressed, pathway enrichment analysis helps us understand what these genes are doing by identifying biological pathways that are overrepresented in a list of differentially expressed genes.
Key Concepts in Pathway Enrichment Analysis
- Biological Pathways: Biological pathways are collections of molecular interactions that govern cellular processes. Examples include signaling pathways (e.g., MAPK pathway) and metabolic pathways (e.g., glycolysis). Pathways are often complex and involve many genes working together.
- Gene Ontology (GO): GO is a framework that provides consistent descriptions of gene products across species. GO terms are categorized into three main domains: biological processes, molecular functions, and cellular components. Pathway enrichment analysis often uses GO terms to interpret the roles of genes.
- Pathway Databases: Several databases store information about known biological pathways, including:
- KEGG (Kyoto Encyclopedia of Genes and Genomes): A database that maps genes to biological pathways.
- Reactome: A curated database of human biological pathways and reactions.
- Biocarta: A repository of biochemical and molecular pathways in human biology.
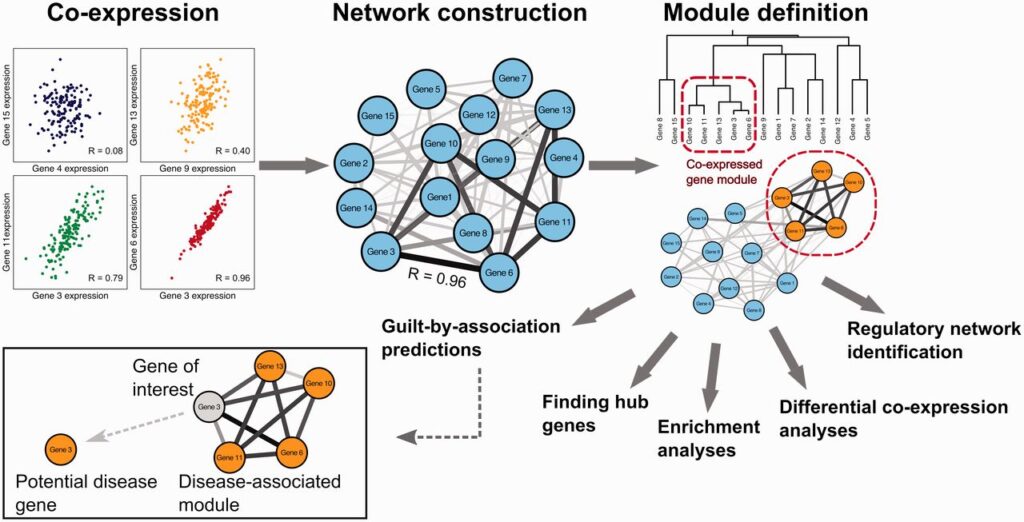
Methods for Gene Expression Profiling and Pathway Enrichment Analysis
Gene Expression Profiling and Pathway enrichment analysis involves comparing a list of differentially expressed genes (identified from gene expression profiling) to known biological pathways to identify which pathways are significantly affected.
- Over-Representation Analysis (ORA): This method compares the number of genes from the input list that are involved in a particular pathway to the expected number based on random chance. If a pathway is significantly overrepresented, it suggests that the pathway is likely involved in the condition being studied.
- Gene Set Enrichment Analysis (GSEA): Unlike ORA, which looks at a predefined list of differentially expressed genes, GSEA analyzes ranked gene expression data. It identifies whether the genes in a particular pathway are disproportionately represented at the top or bottom of the ranked list, indicating their involvement in a biological process.
- Functional Class Scoring (FCS): A more advanced method that aggregates scores for all genes in a pathway to assess whether the pathway, as a whole, is significantly affected.
Workflow of Pathway Enrichment Analysis
- Input Data: The input to a pathway enrichment analysis is typically a list of differentially expressed genes obtained from gene expression profiling experiments.
- Pathway Mapping: Genes are mapped to their respective pathways using databases like KEGG or Reactome.
- Statistical Testing: Statistical tests are performed to determine if the observed overlap between the gene list and the pathways is greater than expected by chance.
- Interpretation of Results: The significantly enriched pathways provide insights into the biological processes affected by the experimental condition.
Applications of Pathway Enrichment Analysis
Pathway enrichment analysis has broad applications across biological research:
- Understanding Disease Mechanisms: By identifying pathways enriched in diseased tissues, researchers can pinpoint the molecular mechanisms underlying conditions like cancer, diabetes, and neurodegenerative diseases.
- Drug Target Identification: Pathways that are significantly altered in response to drug treatments can reveal potential therapeutic targets.
- Biomarker Discovery: Pathway enrichment analysis helps identify key pathways that can serve as biomarkers for disease diagnosis or prognosis.
- Functional Annotation of Gene Lists: It provides biological context for long lists of differentially expressed genes, making it easier to interpret their roles in complex systems.
Integrating Gene Expression Profiling with Pathway Enrichment Analysis
By integrating gene expression profiling with pathway enrichment analysis, researchers gain both a high-level view of which genes are involved in a process and a deeper understanding of how these genes interact within biological pathways. This combination is especially useful in fields such as cancer research, where specific pathways may be dysregulated, leading to uncontrolled cell growth and proliferation.
Case Study: Cancer Research
Imagine a study that aims to identify the molecular changes that occur in lung cancer cells compared to normal lung tissue. Researchers would:
- Perform gene expression profiling: Using RNA-Seq, they measure the expression levels of thousands of genes in both cancerous and healthy tissues.
- Identify differentially expressed genes: They find that several genes are either overexpressed or underexpressed in lung cancer.
- Conduct pathway enrichment analysis: Using these differentially expressed genes, they perform pathway enrichment analysis and discover that the PI3K/AKT signaling pathway, which regulates cell growth and survival, is significantly enriched in the cancer cells.
- Interpret the findings: The results suggest that targeting the PI3K/AKT pathway with specific inhibitors could be a potential therapeutic strategy for treating lung cancer.
Tools for Gene Expression Profiling and Pathway Enrichment Analysis
Several bioinformatics tools and software packages are available to facilitate gene expression profiling and pathway enrichment analysis:
Tools for Gene Expression Profiling:
- DESeq2: A popular R package for analyzing RNA-Seq data and identifying differentially expressed genes.
- EdgeR: Another R package that performs differential expression analysis for RNA-Seq data.
- Cufflinks: A tool that assembles transcripts and quantifies their expression levels from RNA-Seq data.
Tools for Pathway Enrichment Analysis:
- GSEA (Gene Set Enrichment Analysis): A tool that performs GSEA and is commonly used for analyzing gene expression data.
- DAVID (Database for Annotation, Visualization, and Integrated Discovery): A web-based tool that allows researchers to perform ORA and functional annotation of gene lists.
- KEGG Mapper: A tool for mapping genes to KEGG pathways.
Conclusion
Understanding gene expression and how it influences biological pathways is crucial for unraveling the molecular mechanisms underlying health and disease. Gene expression profiling provides detailed insights into the functional state of cells, while pathway enrichment analysis helps researchers interpret this data in the context of known biological pathways. Together, these techniques are transforming research in fields such as cancer biology, drug development, and personalized medicine.
By leveraging gene expression profiling and pathway enrichment analysis, scientists can gain a comprehensive view of how genes and pathways interact, ultimately leading to novel discoveries and therapeutic breakthroughs.
If you want to explore more about applications of Gene Expression Profiling and Pathway Enrichment Analysis you can join us Online for an exciting 2 Day Workshop. More information is available HERE
